| |
|
The APOBEC-1 homolog APOBEC3G acts as an antiviral enzyme.
HIV-1, a human lentivirus, is the major causative agent of AIDS, which presently infects approximately 42 million persons worldwide with 1 million infected persons in North America (http://www.unaids.org). The virus contains a 10-kb single-stranded, positive-sense RNA genome that encodes three major classes of gene products that include: (i) structural proteins such as Gag, Pol and Env; (ii) essential trans-acting proteins (Tat, Rev); and (iii) “auxiliary” proteins that are not required for efficient virus replication in at least some cell culture systems (Vpr, Vif, Vpu, Nef). Among these proteins, Vif is required for efficient virus replication in vivo, as well as in certain host cell types in vitro because of its ability to overcome the action of a cellular antiviral system.
The in vitro replicative phenotype of vif-deleted molecular clones of HIV-1 is strikingly different in vif-permissive cells (e.g. 293T, SUPT1 and CEM-SS T cell lines), as compared to vif-non-permissive cells (e.g. primary T cells, macrophages, or CEM, H9 and HUT78 T cell lines). In the former cells, vif-deleted HIV-1 clones replicate with an efficiency that is essentially identical to that of wild-type virus, whereas in the latter cells, replication of vif-negative HIV-1 mutants is arrested due to a failure to accumulate reverse transcripts and inability to generate infectious proviral integrants in the host cell. These defects are due to the expression, in vif-non-permissive cells, of a host protein called APOBEC3G. The APOBEC3G amino acid sequence exhibits pairwise homology to members of the APOBEC related protein (ARP) family whose functions may be either RNA editing (e.g. APOBEC-1), DNA deamination (e.g. AID) or both. This familial association combined with the known instability of viral reverse transcripts in vif-non-permissive cells in the absence of Vif (and the fact that both Vif and APOBEC3G are present in HIV-1 virions ) strongly suggests that the APOBEC3G antiviral activity may be derived from effects on viral RNA or reverse transcripts. Current research has suggested that HIV RNA/DNA modification is a significant mechanism for the effect of APOBEC3G on viral infectivity.
The role of APOBEC3G as an antiviral enzyme is another active area of research in the Smith lab . The significance of this research is the evaluation of the hypothesis that APOBEC3G deamination activity is essential to antiviral activity and that it will provide a structure-based rationale for this activity and its relationship to Vif. More importantly, the results of these studies should facilitate a rational design of future drugs that target deaminase activity and/or Vif’s ability to overcome endogenous cellular APOBEC3G activity.
|
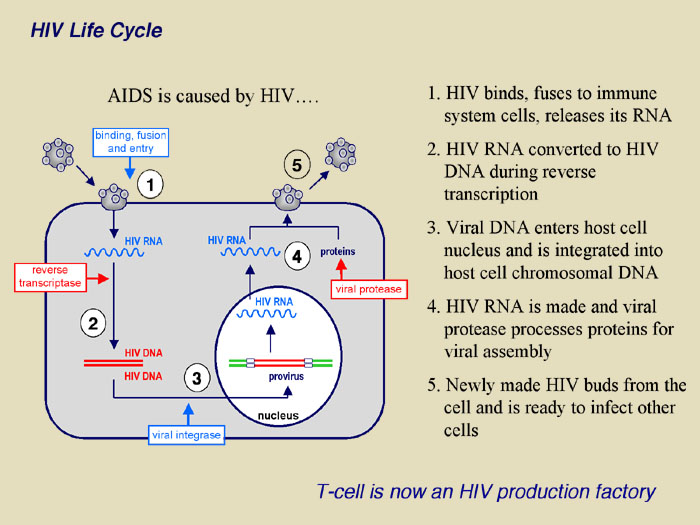
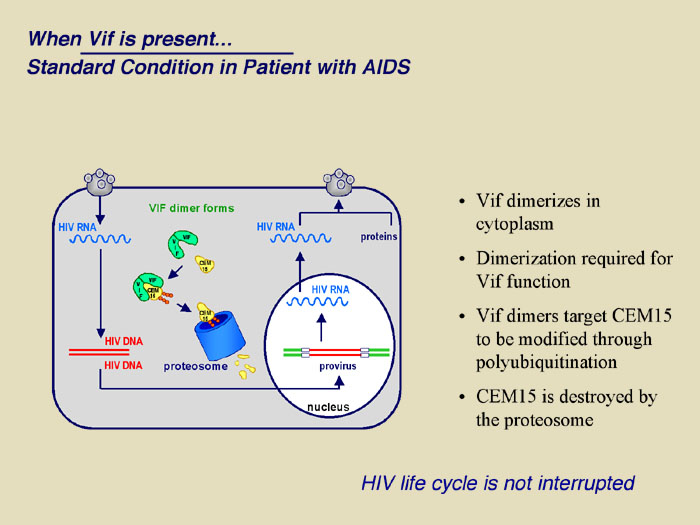
|
The figure demonstrates the recently uncovered mechanism of host cell and HIV-1 interaction that controls viral infectivity and that may be exploited as a novel target for therapeutic intervention. Specifically, the proposed research extends the recent discovery that T cells express an enzyme known as APOBEC3G that functions as a natural defense mechanism against HIV-1 infection. Our current understanding is that APOBEC3G enters viral particles during their assembly and in the subsequent infection, induces dC to dU deamination (editing) at multiple sites throughout the viral genome during and subsequent to minus strand reverse transcription. As such, most or all viral structural and functional proteins may experience simultaneous and multiple mutations leading to a mark reduction in viral infectivity. HIV-1 however, encodes a viral infectivity factor known as Vif that protects HIV-1 from host cell defense by interacting with cellular APOBEC3G, preventing it from entering viral particles and targeting it for degradation.
The relevance of this research to the priorities of HIV/AIDS research is that the findings will address the need for alternative therapeutic approaches. This need is evident in the fact that HIV-1 presently infects approximately 42 million persons worldwide with 1 million infected persons in North America (http://www.unaids.org). This is despite 21 FDA approved therapies. The primary reason AIDS has not been better controlled is due to the high mutation rate of the virus which generates variants of viral proteins that still function in the viral life cycle but do not interact as well or at all with drugs or antibodies. It has been estimated that 16% of new infections in the USA will involve virus that are all ready resistant to approved therapies. Perhaps more concerning is the fact that 17 out of 25 drugs currently in clinical trials are against 'traditional' HIV-1 protein targets, suggesting they may be of limited utility by the time they obtain FDA approval. Therapeutics based on targeting and disrupting the structural constraints necessary for Vif-APOBEC3G interactions, that protect APOBEC3G from Vif binding to it or that activate APOBEC3G are likely to be highly effective because HIV is less likely to be able to produce Vif mutations that escape these drug interactions while at the same time maintaining effective control over APOBEC3G activity.
- Sheehy, A. M., Gaddis, N.C., Choi, J.D. and Malim, M.H. (2002) Nature 418, 646-650.
- Shindo, K., Takaori-Kondo, A., Kobayashi, M., Abudu, A., Fukunaga, K. and Uchiyama, T. (2003) J Biol Chem.
- Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. and Trono, D. (2003) Nature 424, 99-103.
- Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H. and Landau, N. R. (2003) Cell 114, 21-31.
- Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. and Gao, L. (2003) Nature 424, 94-8.
- Lecossier, D., Bouchonnet, F., Clavel, F. and Hance, A. J. (2003) Science 300, 1112.
- Stopak, K., De Noronha, C., Yonemoto, W., and Greene, W.C. (2003) Mol Cell 12, 591-601.
- Yang, B., Gao, L., Li, L., Lu, Z., Fan, X., Patel, C. A., Pomerantz, R. J., DuBois, G. C. and Zhang, H. (2003) J Biol Chem 278, 6596-602.
- Liu, B., Yu, X., Luo, K., Yu, Y. and Yu, X. F. (2004) J Virol 78, 2072-81.
- Sheehy, A. M., Gaddis, N. C. and Malim, M. H. (2003) Nat Med 9, 1404-7.
|
|
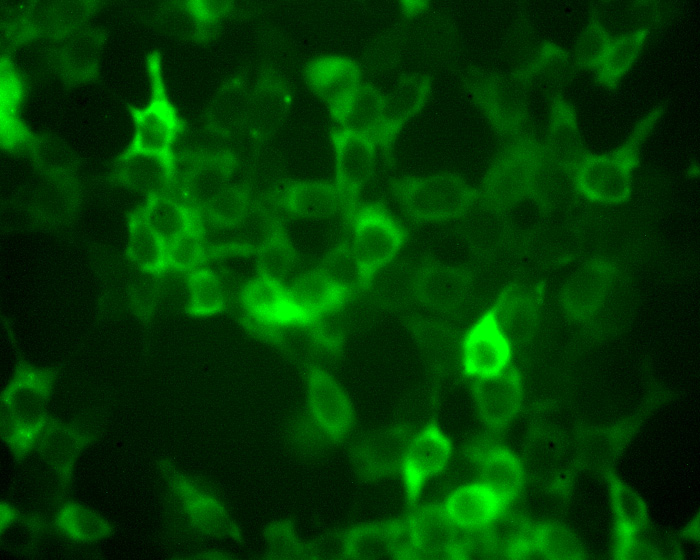
|
The editing enzyme are typically distributed through out the cell, present in both the cytoplasm and nucleus, even though for example apoB mRNA editing only occurs in the nucleus. (return to my homepage to see an image of the subcellular distribution of APOBEC-1 in liver cells). APOBEC3G attacks HIV cDNA during reverse transcription in the cytoplasm. As shown by the image APOBEC3G transfected in 293T cells has a predominantly cytoplasmic distribution.
|
|
|
|