|
RNA Editing and DNA Mutagenesis
The Smith Lab investigates a family of enzymes known as APOBEC-1 related proteins (ARPs) whose function is to change the nucleotide sequence of RNA or DNA. The consequence of this change frequently is an alteration in protein coding capacity and hence ARPs are thought to bring about diversity in the proteome of cells. It might seem obvious that such 'power' would need to be precisely 'tuned' so that it only affected select nucleic acids sequences, in certain cells at vary particular times. In fact, all experimental data to date suggest that enzyme expression and activity are highly regulated.
In recent years researchers have become increasing aware of the complexity of processes leading to cell and tissue function. The sequencing of genomes has now established that the linear sequence of DNA that apparently is dedicated to protein coding is one to two orders of magnitude below that which one might predict to exist given the diversity of proteins expressed in cells. Mutation and recombination of DNA, use of alternative promoters, alternative pre-mRNA splicing, alternative polyadenylation and mRNA turnover contribute to the diversity of cellular RNA and correspondingly, to the vastly greater number of expressed proteins than is apparent from genomic coding capacity. Using predictive algorithms, bioinformatics has posited that >60% of human primary transcripts are alternatively spliced; suggesting that alternative mRNA splicing may be the rule rather than the exception. RNA editing can also significantly diversify the proteome. Although, relatively few editing events occur in mammals they have profound affects on the function of transmembrane receptors and ion channels, in erythropoiesis and inflammation in cardiovascular disease in cancer and upon the life cycle of viruses. The family of mammalian ARPs also have activity on DNA and it is quite evident their site-selective mutation of DNA through deoxycytidine deamination to deoxyuridine (DNA editing) is regulated by cells to enable diverse protein expression for the genome or prevent protein expression from viruses (see AID and APOBEC3G descriptions below).
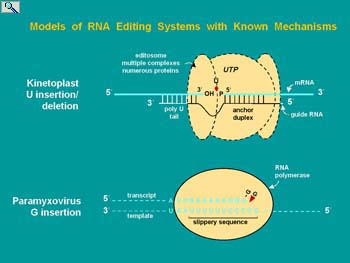
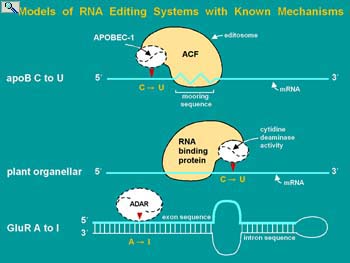
The Smith lab entered this field at a time when very little was known but the discovery of RNA editing had clearly identified this form of gene regulation as a burgeoning area of research. Several types of mRNA editing were discovered simultaneously or with a year of each other. Protozoan guide RNA-dependent mRNA editing was responsible for the assembly of the majority of mRNAs encoded by the mitochondria. The genome codes for incoherent segments of these mRNAs (known as cryptogenes) and the gRNAs required for editing the primary transcripts to functional mRNAs. As a more discrete process, the glutamate-gated calcium channel subunits of the mammalian central nervous system acquire appropriate charge to regulate calcium flux by site-specific, adenosine to inosine (base pairs like guanosine) mRNA editing. The Adenosine Deaminase Active on RNA (ADARs) responsible for this mRNA editing are expressed in all eukaryotes and have numerous other mRNA substrates. Plant mitochondria and chloroplast mRNAs undergo extensive cytidine to uridine deamination affecting the coding potential of numerous mRNAs and the diversity of proteins that can be expressed. In fact the editing of plant organellar mRNAs is so extensive that prior to the discovery of mRNA editing it was not apparent which genes coded for proteins.
|
The entry point for the Smith lab was in the area of cytidine to uridine apolipoprotein B mRNA editing. This form of editing converts a CAA glutamine codon to a UAA stop codon, enabling a long and a short form of apoB protein to be expressed. Both proteins served in the transport of triglycerides and cholesterol in blood but the expression of the longer form is associated with an elevated risk of atherosclerosis leading to cardiovascular disease. For more background on RNA editing please see:
The specific focus of the research is to identify and characterize novel mammalian mRNA editing systems that employ a zinc-dependent deamination mechanism for the post-transcriptional conversion of cytidine to uridine at select sites within mRNAs. Computational modeling has suggested a family of mammalian enzymes known as ARPs as responsible for C to U editing of nucleic acids. The expression of these enzymes in biology suggests that mRNA editing and DNA mutagenesis may be involved in numerous physiological processes and could be manipulated for the prevention of cardiovascular disease, HIV infection and cancer, and is also necessary for production of antibodies in B lymphocytes. Recent evidence indicates that the enzyme involved in suppressing HIV-1 infectivity (APOBEC3G) and the enzyme that promotes antibody production (AID) may act to mutate deoxy cytidine in DNA rather than or in addition to RNA. Our research involves molecular biology and protein techniques, DNA microarray analyses and computational biology to identify the mRNAs that are edited by CDARs and to determine the biological consequence of these editing events in terms of the predicted changes in the types of protein structures and functions that can be expressed. Our studies have demonstrated how cytidine to uridine mRNA editing contributes to expansion in the diversity of expressed mRNA sequences known collectively as the transcriptome. We are also evaluating the regulatory mechanisms controlling the expression of editing factors and their localization in the cell nucleus. The development of this new information will establish an important new annotation of the human genome that will serve as a frame of reference for studies of proteins involved in health and disease and mechanisms regulating their expression.
|