| |

|
|

|
|
|
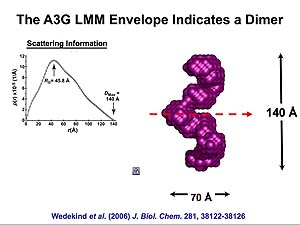
Molecular envelop calculated from data obtained by Small Angle X-ray Scattering (SAXS) for RNA-depleted, catalytically active human APOBEC3G expressed in, and purified from Sf9 cells. The molecular envelop suggest that catalytically active APOBEC3G is a homodimer with intersubunit contacts within the C-termini. From: Wedekind, et al., J. Biol. Chem. 281, 38122-38126 (2006). Animations prepared courtesy of Dr. J.E. Wedekind.
|
|
|
|
|
|
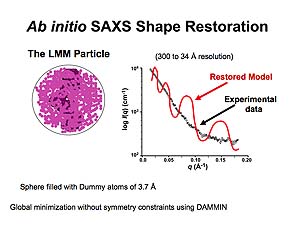
Sphere encompassing all of the collected data is filled with dummy atoms that are progressively eliminated through global minimization to achieve the molecular volume occupied by APOBEC3G. Animations prepared courtesy of Dr. J.E. Wedekind.
|
|
|
Through numerous iterations of this process, the restored model is determined with the best fit to the experimental data.
|
|
|

|
|
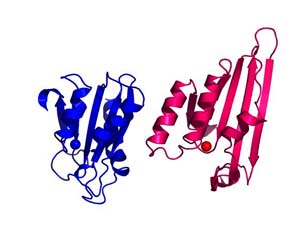
Alignment of the zinc-dependent cytidine deaminase domains characterized in the crystal structures from the yeast RNA and DNA editing enzyme Cdd1 with that found in APOBEC2. The domain has deaminase activity in Cdd1 but does not in APOBEC2; suggesting that additional structural considers are relevant in understanding activity of the APOBEC family. Animations prepared courtesy of Dr. J.E. Wedekind.
|
|
|
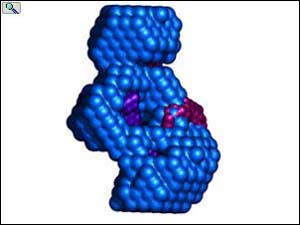
Transparent surface and ribbon depiction of the CDD1 cytidine deaminase structure (Protein DataBank entry 1R5T); each CDA subunit is colored differently and contains a single ZDD signature motif. Inset, a single CDA domain (residues 1-132) representing the fundamental / fold; Zn2 is depicted as a gray sphere. Right, the hA3G-DR (LMM) envelope (red, semitransparent) with two independent CDA domains of CDD1 docked inside. From: Wedekind, et al. J. Biol. Chem. 281, 38122-38126 (2006). Animations prepared courtesy of Dr. J.E. Wedekind.
|
|
|
|
|
APOBEC3G bound to RNA to cellular RNA has a minimal size of a tetramer. The molecular volume of the tetramer has been determined by Small Angle X-ray Scattering. From: Wedekind, et al. J. Biol. Chem. 281, 38122-38126 (2006). Animations prepared courtesy of Dr. J.E. Wedekind.
|
|
|
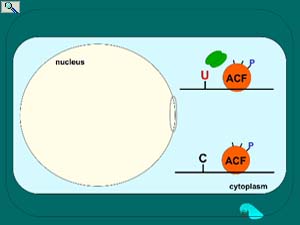
Regulation of apoB mRNA editing through phosphorylation of the RNA-binding and APOBEC1-binding protein known as ACF. ACF is predominantly cytoplasmic (see fluorescence microscopy and electron microscopy below) but upon insulin or ethanol stimulation of the liver, trafficks to the nucleus. Once in the nucleus, protein kinase C phosphorylates select serines on ACF to achieve tight binding to APOBEC1 and activation of editing activity. Protein phosphatase 1 removes these high turnover phosphates as what is hypothesized to be a requirement for ACF to traffick back to the cytoplasm. Nuclear export of ACF is proposed facilitate apoB mRNA nuclear export. Smith, et al. In "Topics in Current Genetics" (Grosjean, H., ed), Springer-Verlag, pp. 1610-2096 (2005); Lehmann, et al. Nucleic Acids Res. 34: 3299-3308 (2006); Lehmann, et al. FBiochem. Biophys. Acta. 1773:408-418 (2007). Animation prepared by Jenny M.L. Smith.
|
|
|
|
|
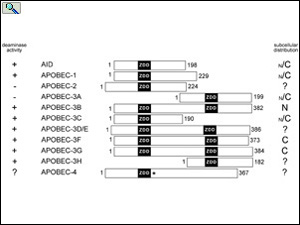
Alignment of the APOBEC family members draw to scale showing the relative positions of the zinc-dependent deaminase domains (ZDD) in each protein. Subcellular localization of each APOBEC protein is indicated to the right. From: Smith, The APOBEC1 Paradigm for Mammalian Cytidine Deaminases that Edit DNA and RNA. in RNA and DNA Editing (H. Grosjean, ed.) Chapter 3, Landes BioScience, TX (2009).
|
|
|
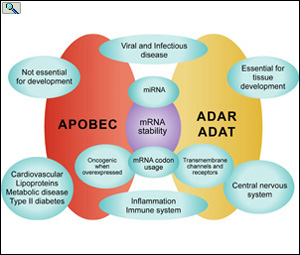
Function of APOBEC, ADAR and ADAT editing proteins in biological systems. From: From: Smith, H.C. The APOBEC1 Paradigm for Mammalian Cytidine Deaminases that Edit DNA and RNA. in RNA and DNA Editing (H. Grosjean, ed.) Chapter 3, Landes BioScience, TX (2009).
|
|
| |
|
|
| |
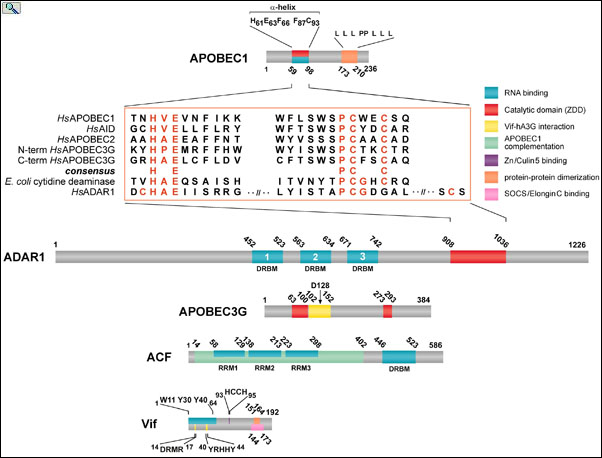
Alignment of conserved residues (red) with the ZDD of APOBEC and ADRA editing enzymes. APOBEC-1, ADAR1, APOBEC3G, APOBEC1 Complementation Factor (ACF) and HIV Viral Infectivity Factor (Vif) are drawn to scale with their relevant functional domains indicated. ACF binds APOBEC1 and binds to RNA editing substrates enabling site-specific C to U RNA editing. Vif binds to APOBEC3G and chaperones it to the proteosome for degradation, thereby removing APOBEC3G host-defense function from the cell. From: Smith, The APOBEC1 Paradigm for Mammalian Cytidine Deaminases that Edit DNA and RNA. in RNA and DNA Editing (H. Grosjean, ed.) Chapter 3, Landes BioScience, TX (2009).
|
|
| |
|
|
| |
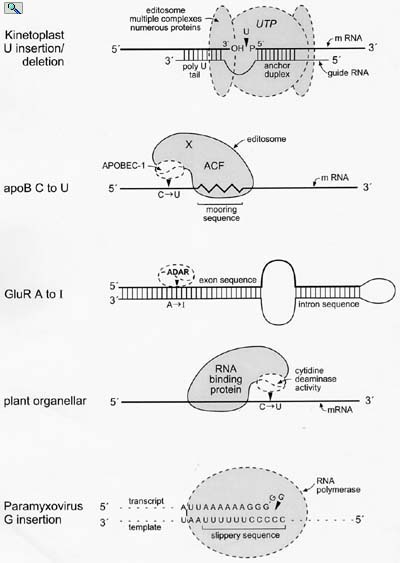
Models for the minimal editing complexes involved in known RNA editing activities in eukaryotic cells. From: Wedekind, et al. Trends in Genetics 19:207-216 (2003).
|
|


|
|